磷脂纳米盘Nanodisc的产品应用

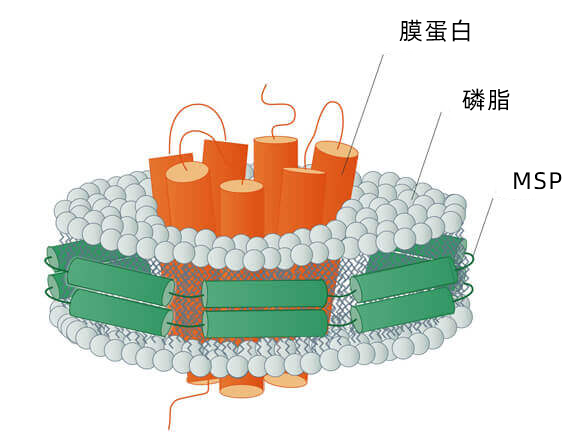
Nanodisc纳米盘的用途
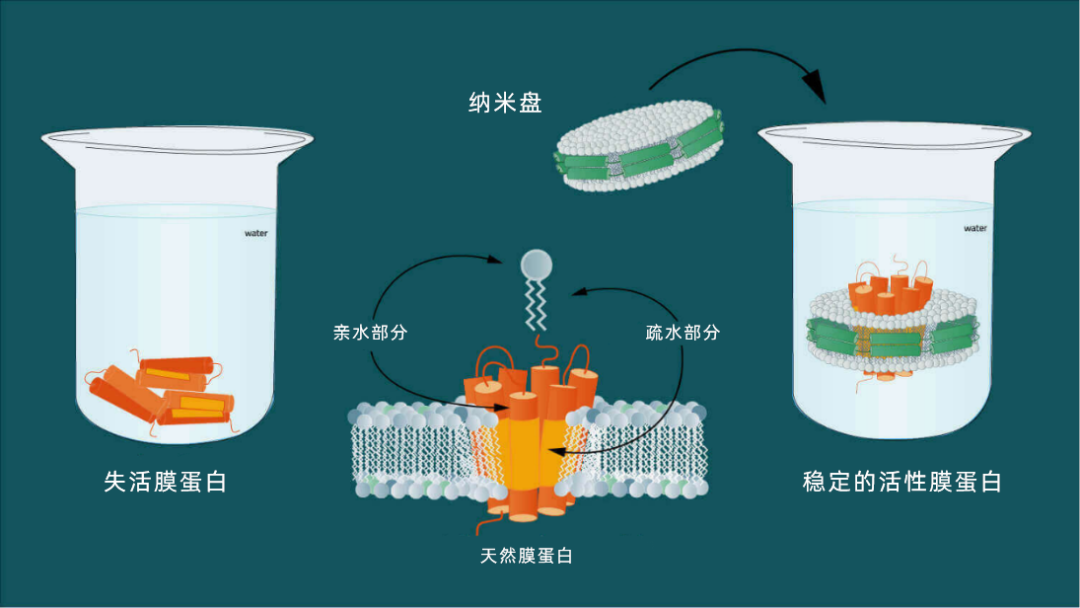
图2:膜蛋白同时具有疏水性和亲水性。纳米盘可以使膜蛋白溶于水溶液。
纳米盘用于模拟细胞的天然磷脂双分子层,以此来寻找靶分子(通常为膜蛋白)。膜蛋白是细胞间通讯的关键物质,介导基本的生物过程,如信号转导、跨膜转运过程、化学信号传感及胞间相互作用的协调。
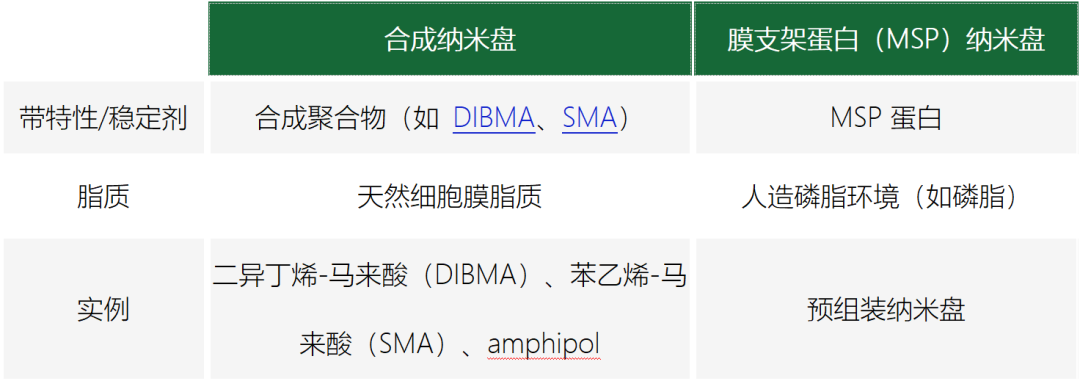
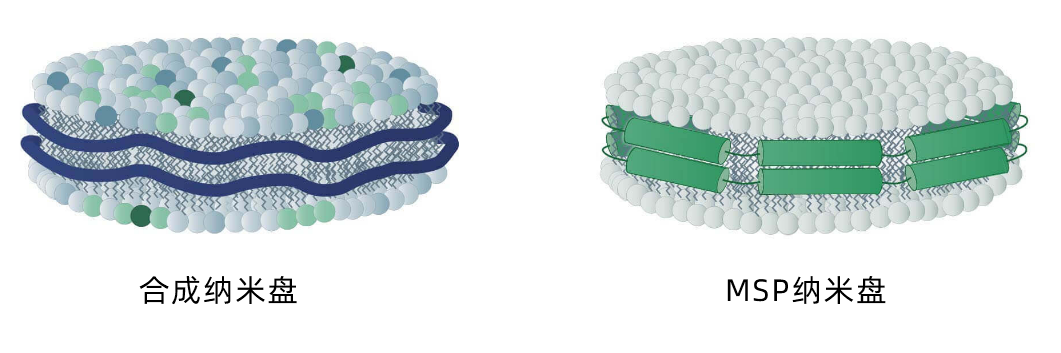
但首先,我们将分别介绍这两种纳米盘,先从 MSP 纳米盘开始。
MSP纳米盘
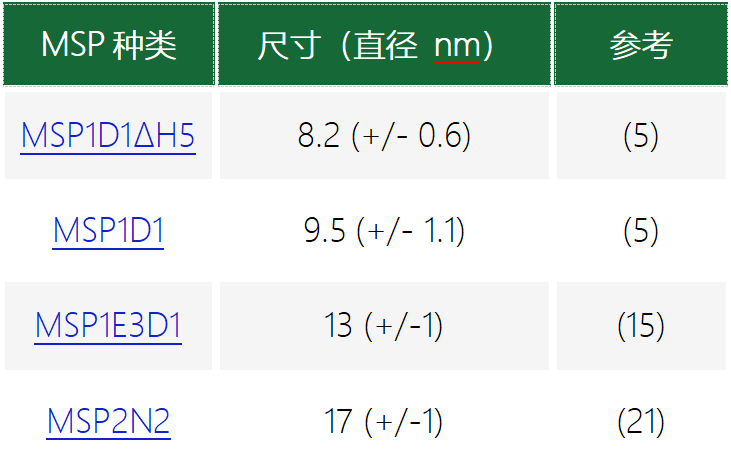
MSP纳米盘的其他优点
与其他膜蛋白增溶和重组系统相比,MSP纳米盘具备许多优势,尤其在配体结合研究、构象动力学分析以及蛋白质相互作用研究等方面(6)。纳米盘可用于在类似于天然膜的人工环境中重组GPCR 或转运蛋白等膜蛋白质。

这些经由纳米盘稳定的蛋白质可通过常规的色谱层析程序直接纯化。得到的纯化膜蛋白-纳米盘复合物可应用于需要接触蛋白质的生理细胞内及细胞外表面的场景,从而能够不受限制地接触拮抗剂、激动剂、G 蛋白及其他相互作用对象(7)。
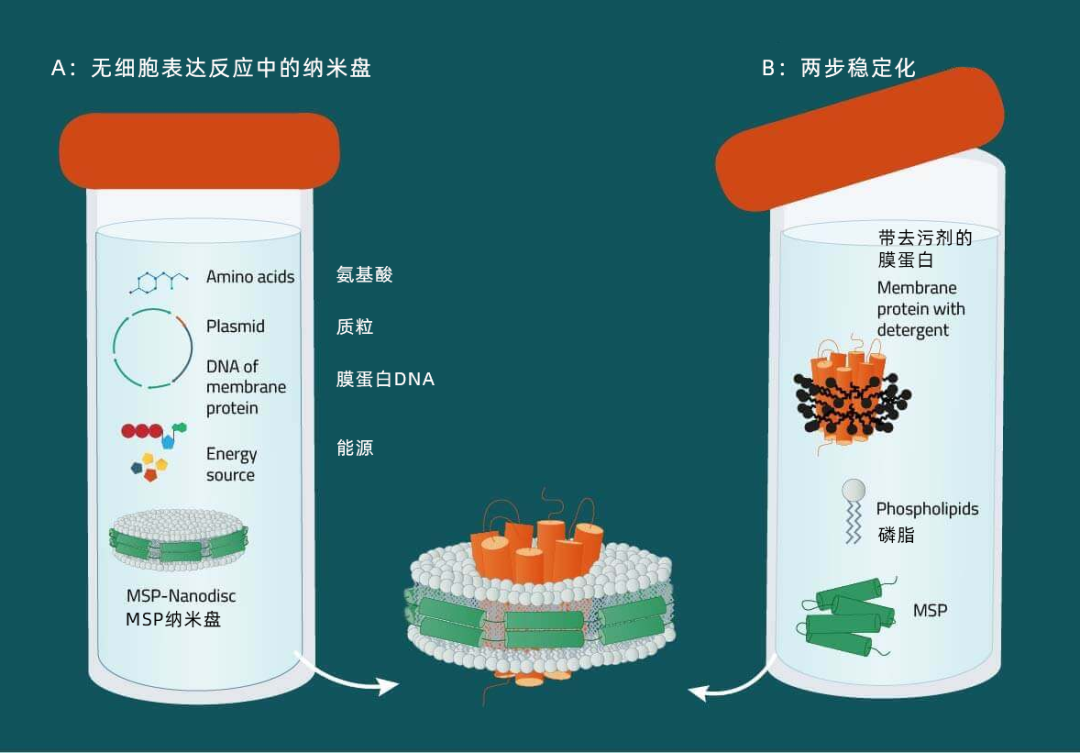
A:结合纳米盘与无细胞表达系统从表达质粒开始,可在无细胞系统中生成膜蛋白。在整合了初生膜蛋白(8)的混合物中提供预组装纳米盘。不需要额外添加去污剂,这样可以最大限度地减少伪影的存在。作为一个可选项,可以将生物素酰化或同位素标记等修饰包含在内。
B:去污剂增溶蛋白质的两步重组从合适去污剂中的纯化膜蛋白开始,添加膜支架蛋白和磷脂。包含膜蛋白的纳米盘可自发形成,并通过亲和或排阻层析法纯化得到(6,7)。
C:直接从膜增溶从表达目标蛋白的膜开始,添加去污剂和膜支架蛋白(MSP)。膜磷脂、膜蛋白和 MSP 组装形成纳米圆盘复合物(5)。此处,获得了表征膜蛋白群的纳米盘复合物的混合物,可用于蛋白质组学研究。如果需要,可采用亲和色谱法纯化对单个膜蛋白-纳米盘复合物进行纯化处理。与方法 B 相比,接触去污剂的时间明显缩短了(从按天数缩短为按小时)。
磷脂的选择——维持适当蛋白质活性的关键
如前所述,MSP 纳米盘的磷脂成分是人造的。这意味着,必须事先确定好构成目的膜蛋白所需人工膜环境的磷脂。但存在多种磷脂可供选择,应该选择哪一种呢?
对于这个问题,请参阅我们最常用于 MSP 纳米盘的磷脂列表:
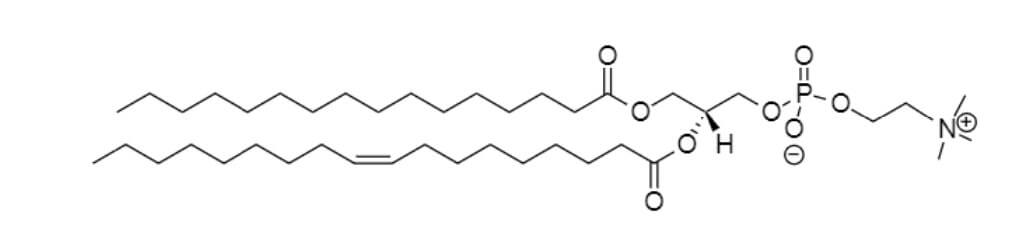
二肉豆蔻酰磷脂酰胆碱(DMPC)
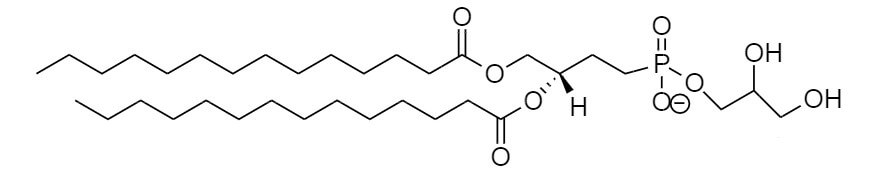
棕榈酰-油酰-磷脂酰胆碱(POPC)
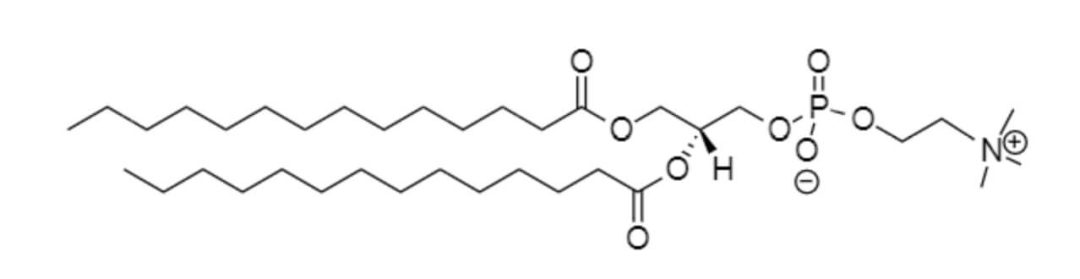
磷脂酰甘油(DMPG)
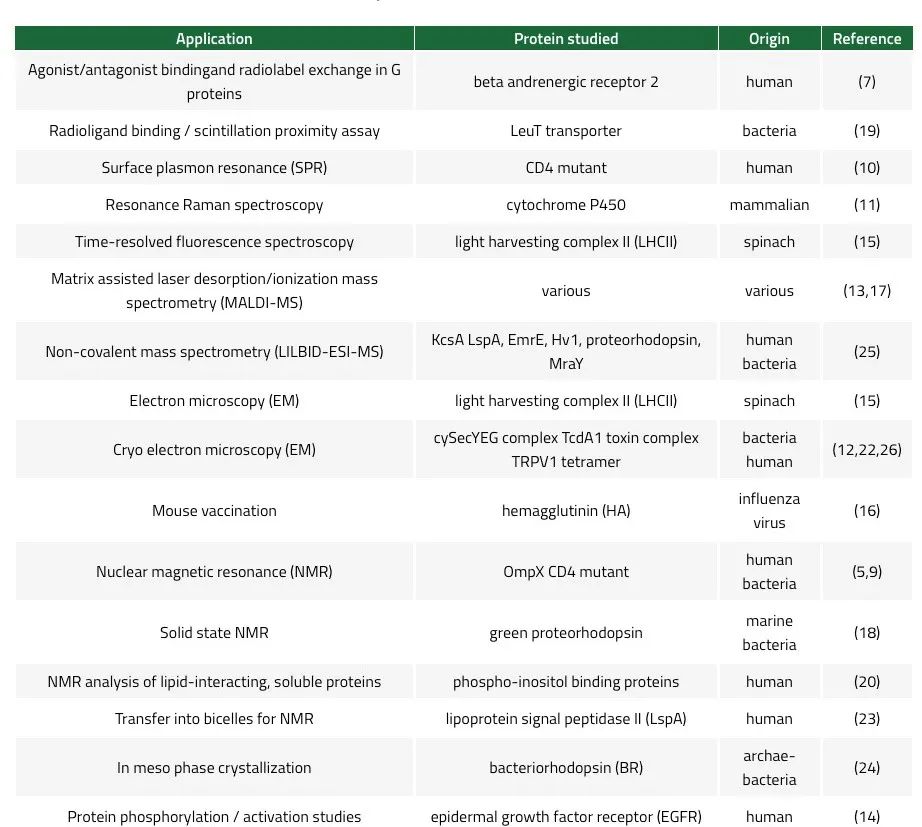
纳米盘在科学中的应用实例
MSP纳米盘最初是由Sligar及其同事一起发表的(3,4)。MSP纳米盘为稳定膜蛋白提供了完美的环境,使得通过NMR 和 SPR(9,10)等方法研究配体、激动剂或拮抗剂的结合成为可能。纳米盘被证实可以提高 在冷冻电镜中跨膜蛋白区的分辨率(22,26)。膜支架蛋白可采用组氨酸标记,以促进蛋白质纳米盘复合物的纯化、检测和固相处理。其他纳米盘应用的实例包括共振拉曼(11)、MALDI(13)、非共价质谱法(25)、蛋白激活研究(14)、时间分辨荧光光谱(15)和蛋白质结晶(24)。重组成纳米盘的抗原已被用于提高小鼠的免疫原应答,显示出其作为疫苗的潜力 (16)。此外,“大肠杆菌”的整个膜蛋白质组被重组为纳米盘,从而创建出水溶性膜蛋白库 (15)。在纳米盘中重组的蛋白质可以转移到双层膜微胞中以提高 NMR 分辨率(23)。在纳米盘的协助下,甚至实现了对可溶性蛋白质以及与脂质相互作用的蛋白质的分析 (20)。表 3 列出了纳米盘应用的实例。
参考文献
1. Douglas, Shawn M., James J. Chou, and William M. Shih. "DNA-nanotube-induced alignment of membrane proteins for NMR structure determination." Proceedings of the National Academy of Sciences 104.16 (2007): 6644-6648.
2. Yeates, T. O., et al. "Structure of the reaction center from Rhodobacter sphaeroides R-26: membrane-protein interactions." Proceedings of the National Academy of Sciences 84.18 (1987): 6438-6442.
3.Bayburt, T.H. et al. Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J. Struct. Biol. (1998), 123(1):37-44
4.Civjan, N.R. et al. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. BioTechniques (2003) 35:556-563
5.Hagn, F. et al. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J.Am.Chem. Soc. (2013), 135:1919-1925
6.Serebryany et al. Artificial membrane-like environments for in vitro studies of purified G-protein coupled receptors. Biochim. Biophys. Acta (2012), 181:225-233
7.Leitz, J. et al. Functional reconstitution of beta2-adrenergic receptors utilizing self-assembling nanodisc technology. BioTechniques (2006), 40:601-612
8.Proverbio D., et al. Functional properties of cell-free expressed human endothelin A and endothelin B receptors in artifical membrane environments. Biochim.Biophys. Acta (2013), 1828(9):2182-92
9.Glueck, J.M. et al. Integral membrane proteins in nanodiscs can be studied by solution NMR spectroscopy. J.Am.Chem.Soc. (2009), 131(34):12060-1
10. Glueck, J.M. et al. Nanodiscs allow the use of integral membrane proteins as analytes in surface plasmon resonance studies. Anal. Biochem. (2011), 408(1):46-52
11.Mak, P.J. et al. Defining CYP3A4 structural responses to substrate binding. Raman spectroscopic studies of a nanodisc-incorporated mammalian cytochrome P450. J.Am.Chem.Soc. (2011) 133(5):1357-66
12. Frauenfeld, J. et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nature Struct. Mol. Biol. (2011), 5:614-21
13.Marty M.T., et al. Ultra-thin layer MALDI mass spectrometry of membrane proteins in nanodiscs. Anal. Bioanal. Chem. (2012) 402(2):721-9
14.Wang, Z. et al. Tyrosine phosphorylation of Mig6 reduces its inhibition of the epidermal growth factor receptor. ACS Chem. Biol. (2013) 8(11):2372-6.
15.15. Pandit A., et al. Assembly of the major light-harvesting complex II in lipid nanodiscs. Biophys. J. (2011) 101:2507-2515
16. Bhattacharya, P. et al. Nanodisc-incorporated hemagglutinin provides protective immunity against influenza virus infection. J. Virology (2010) 361-371
17.Marty M.T. et al., Nanodisc-solubilized membrane protein library reflects the membrane proteome. Anal. Bioanal. Chem. (2013) 405(12):4009-16
18.Moers et al., Modified lipid and protein dynamics in nanodiscs. Biochim. Biophys. Acta (2013), 1828(4):1222-9.
19.Nasr. et al., Radioligand binding to nanodisc-reconstituted membrane transporters assessed by the scintillation proximity assay. Biochemistry (2014), 14;53(1):4-6.
20.Kobashigawa. et al., Phosphoinositide-incorporated lipid-protein nanodiscs: A tool for studying protein-lipid interactions. Anal. Biochem. 410 (2011), 77-83
21.Grinkova, Y.V., et. al., Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Engineering, Design & Selection (2010) 23(11):843-848
22.Gatsogiannis, C, et. al., Membrane insertion of a Tc toxin in near-atomic detail. Nat. Struct. Mol Biol. (2016) Oct;23(10):884-890.
23.Laguerre, A. et al. From nanodiscs to isotropic bicelles: A procedure for solution nuclear magnetic resonance studies of detergent-sensitive integral membrane proteins. Structure (2016) 24, 1-12.
24.Nikolaev, M. et al. Integral membrane proteins can be crystallized directly from nanodiscs. Cryst. Growth Des. (2017) 17(3), 945–948
25.Henrich, E., et al. Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology. eLife (2017) 6:e20954.
26.Gao, Y. et al. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature (2016) 534(7607):347-351. doi:10.1038/nature17964

















 首页
首页 Facebook
Facebook WeChat
WeChat Twiitter
Twiitter
